Non-negative Matrix Factorization for Time-Resolved Raman Spectroscopy Data: Difference between revisions
No edit summary |
m Correcting the \Sigma symbol used in the definition of SVD |
||
| Line 21: | Line 21: | ||
<b><u>Pre-proceesing</u></b> | <b><u>Pre-proceesing</u></b> | ||
Singular Value Decomposition (SVD) for primary factorization <math> \mathbf{M^{T}} =</math> '''U''' <math> \ | Singular Value Decomposition (SVD) for primary factorization <math> \mathbf{M^{T}} =</math> '''U''' <math> \mathbf \Sigma \mathbf{V^{T}} </math>. | ||
Generate <math> \mathbf{U} </math> from '''U''' with <math> \mathbf{U}[:,0] = [ 1 , 1 , ... , 1 , 1 ]^T </math> and <math> \mathbf{U}[ : , 1 : ] = </math> '''U''' <math> [ : , : r - 1 ] </math>. | Generate <math> \mathbf{U} </math> from '''U''' with <math> \mathbf{U}[:,0] = [ 1 , 1 , ... , 1 , 1 ]^T </math> and <math> \mathbf{U}[ : , 1 : ] = </math> '''U''' <math> [ : , : r - 1 ] </math>. | ||
Revision as of 18:34, 29 November 2022
PID (if applicable): doi:10.1007/s10910-020-01201-7
Problem Statement
Crystallization of Paracetamol in Ethanol
Object of Research and Objective
Determination of Intermediate States and their Kinetics along the Crystallization of Paracetamol in Ethanol using Raman Spectroscopy.
Procedure
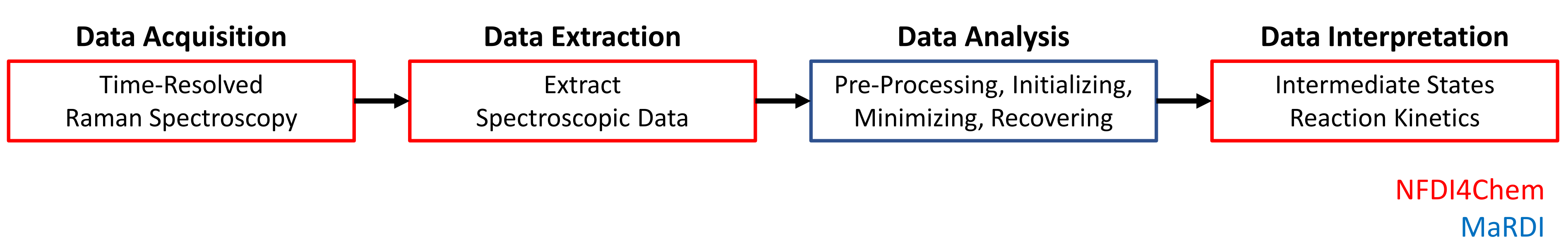
1. Data Acquisition
Time-resolved Raman Spectroscopy to follow the crystallization of Paracetamol in accoustically-levitated Ethanol droplets. Surface, temperature (22.0 +/- 1.0 °C), and relative humidity (17.5 +/- 2.5 %) of the environment are controlled by Nitrogen stream.
2. Data Extraction
Extract the spectroscopic measurement matrix ( measurements of non-negative intensities) from measurement file containing spectroscopic data and metadata.
3. Data Analysis
Factorize such that ( with the rank of factorization or expected number of components) using a novel non-negative matrix factorization (NMF) approach:
Pre-proceesing
Singular Value Decomposition (SVD) for primary factorization U .
Ensure orthogonality among columns of .
Initializing
Apply PCCA+ to to obtain the transformation matrix to initialize , , and .
( the pseudoinverse of singular / non-square matrices)
( defined as without the first / last row)
Minimizing
Objective function maintains positivity (1,2,4), column (3) and row stochastics (5), depending on , , and .
Minimization of with respect to adjusts and numerically to the claimed structural properties.
Recovering
Minimization returns , which allows to recover , , and .
4. Data Interpretation
contains the spectra of the substances involved in the crystallization process, while allows an inference on the kinetics. Interpretation of both matrices leads to the identification of intermediate states and the underlying kinetics.
Involved Disciplines
NFDI4Chem (wikidata:Q96678459)
Raman Spectroscopy (wikidata:Q862228)
MaRDI (wikidata:Q108327788)
Numerical Analysis (wikidata:Q11216)
Mathematical Optimization (wikidata:Q141495)
Linear Algebra (wikidata:Q82571)
Data Streams
NFDI4Chem MaRDI (.txt File containing measurement matrix )
MaRDI NFDI4Chem (.png Files containing component spectra and relative concentration profiles )
Model
A measurement matrix is factorized in a matrix , containing the spectra of the substances involved, and a matrix , which allows an inference on the kinetics.
Discretization
- Time: Time-resolution of Raman Spectroscopy
- Space: -
Variables
Parameter
| Name | Unit | Symbol |
|---|---|---|
| Temperature | °C | T |
| Relative Humidity | % | RH |
| Rank of Factorization | - | |
| Number of Singular Values | - | |
| Singular Value Tolerance | - | |
| Objective Function Parameter | - | |
| Maximum Number of Iterations | - |
Process Informationen
Process Steps
Applied Methods
| ID | Name | Process Step | Parameter | realised/implemented by |
|---|---|---|---|---|
| doi:10.1007/s10910-020-01201-7 | NMF Algorithm | Data Analysis | Matlab Script | |
| wikidata:Q420904 | SVD | Data Analysis - Pre-Processing | Matlab R2019a | |
| wikidata:Q43219517 | Pseudoinverse | Data Analysis - Initializing | Matlab R2019a | |
| doi:10.1016/j.laa.2004.10.026 | PCCA+ | Data Analysis - Initializing | Matlab Script | |
| wikidata:Q1253278 | Nelder-Mead Algorithm | Data Analysis - Minimizing | Matlab R2019a | |
| wikidata:Q43219517 | Pseudoinverse | Data Analysis - Recovering | Matlab R2019a |
Software used
| ID | Name | Description | Version | Programming Language | Dependencies | versioned | published | documented |
|---|---|---|---|---|---|---|---|---|
| - | iC Raman | Data Acquisition & Reaction Analysis | 4.1 | ? | Windows | Yes | Yes | Yes |
| sw:558 | Matlab | Programming and numeric Computing | R2019a | C,C++,Fortran,Java | Windows, Mac, Linux | Yes | Yes | Yes |
Experimental Devices/Instruments and Computer-Hardware
| ID | Name | Description | Version | Part Nr | Serial Nr | Location | Software |
|---|---|---|---|---|---|---|---|
| - | Raman RXN1 | Spectrometer | |||||
| - | GenunineIntel | Intel(R) Core(TM) i7-9700T CPU @ 2.00 GHz |
Input Data
Output Data
Reproducibility
Reproducibility of the Experiments on the original Devices/Instruments/Hardware
Yes
Reproducibility of the Experiments on other Devices/Instruments/Hardware
Yes
Transferability of the Experiments to
a) other solutes, solvents, parameter
b) other chemical reactions
Legend
The following abbreviations are used in the document to indicate/resolve IDs:
doi: https://dx.doi.org/
sw: https://swmath.org/software/
wikidata: https://www.wikidata.org/wiki/










![{\displaystyle \mathbf{U}[:,0] = [ 1 , 1 , ... , 1 , 1 ]^T }](https://wikimedia.org/api/rest_v1/media/math/render/svg/26709c60d988851e25cde4f9f262c5fc5adbfc32)
![{\displaystyle \mathbf{U}[ : , 1 : ] = }](https://wikimedia.org/api/rest_v1/media/math/render/svg/82afc9de4698797ec004fc06b32d49c39a102e49)
![{\displaystyle [ : , : r - 1 ] }](https://wikimedia.org/api/rest_v1/media/math/render/svg/007da38dab9ff251660dc82fb4256a318b40c51b)




























