Portal/TA2: Difference between revisions
m →Interns and Student Assistants: add Saksham's info |
→Ongoing initiatives: Changed links to mathalgodb. |
||
| (12 intermediate revisions by 3 users not shown) | |||
| Line 7: | Line 7: | ||
==Ongoing initiatives== | ==Ongoing initiatives== | ||
===AlgoData=== | ===MathAlgoDB (formerly AlgoData)=== | ||
Our goal is to establish a knowledge graph of numerical algorithms, which interlinks those algorithms with the addressed mathematical problems and associated research data such as journal papers or implementing software packages. As often many variations of essentially the same numerical algorithm are discussed in the literature, an editorial board of domain experts will define the individual algorithm items in the database. The interlinking with the associated data, however, is carried out semi-automatically, using various data sources such as text-mining or metadata search in other databases and MaRDI services. | Our goal is to establish a knowledge graph of numerical algorithms, which interlinks those algorithms with the addressed mathematical problems and associated research data such as journal papers or implementing software packages. As often many variations of essentially the same numerical algorithm are discussed in the literature, an editorial board of domain experts will define the individual algorithm items in the database. The interlinking with the associated data, however, is carried out semi-automatically, using various data sources such as text-mining or metadata search in other databases and MaRDI services. | ||
The measure improves the findability of research data associated with given numerical algorithms and allows non-experts, who need to solve numerical problems in their scientific work, to quickly gain an overview of available methods, their characteristics and implementations. Mathematicians in scientific computing are empowered to easily track the field’s progress by data on competing methods. | The measure improves the findability of research data associated with given numerical algorithms and allows non-experts, who need to solve numerical problems in their scientific work, to quickly gain an overview of available methods, their characteristics and implementations. Mathematicians in scientific computing are empowered to easily track the field’s progress by data on competing methods. | ||
We provide an interface for querying and editing data. Note that editing and proposing data is not fully functional yet. The interface comes in a stable version and a beta version which reflects our current working status. The permanent links are [https:// | We provide an interface for querying and editing data. Note that editing and proposing data is not fully functional yet. The interface comes in a stable version and a beta version which reflects our current working status. The permanent links are [https://mathalgodb.mardi4nfdi.de mathalgodb.mardi4nfdi.de] and [https://main.m1.mardi.ovh main.m1.mardi.ovh]. | ||
The beta version is updated daily. Use the deep links below to access Data and the Ontology definition. | The beta version is updated daily. Use the deep links below to access Data and the Ontology definition. | ||
| Line 22: | Line 22: | ||
|- | |- | ||
|Ontology Link | |Ontology Link | ||
|[https:// | |[https://mathalgodb.mardi4nfdi.de/static/widoco/v1/index-en.html Ontology] | ||
|[https://main.m1.mardi.ovh/static/widoco/v1/index-en | |[https://main.m1.mardi.ovh/static/widoco/v1/index-en.html Ontology] | ||
|- | |- | ||
|Simple Search | |Simple Search | ||
|[https:// | |[https://mathalgodb.mardi4nfdi.de/ Search] | ||
|[https://main.m1.mardi.ovh/ Search] | |[https://main.m1.mardi.ovh/ Search] | ||
|- | |- | ||
|Graph Search | |Graph Search | ||
|[https:// | |[https://mathalgodb.mardi4nfdi.de/search/advanced/ Search] | ||
|[https://main.m1.mardi.ovh/search/advanced/ Search] | |[https://main.m1.mardi.ovh/search/advanced/ Search] | ||
|} | |} | ||
| Line 36: | Line 36: | ||
The ontology definition in standard formats (XML, Triples, Turtle) is available under the ontology link above. | The ontology definition in standard formats (XML, Triples, Turtle) is available under the ontology link above. | ||
At this point, we accept submissions for the graph, respecting the ontology, in standard formats. Please use the definition of the sample Model Order Reduction-Graph as a template. We also support a customized meta format which permits a simpler definition of the graph and has DOI search capabilities for bulk additions to the graph. | |||
===Open Interfaces=== | |||
The scientific-computing community uses several different programming languages for implementing numerical solvers. Moreover, these solvers often have significantly different programming interfaces. Both of these factors impede interoperability, as switching from, say, a Python solver for time integration to another solver implemented in C requires writing bindings and adapting the calling code to different function names and the order of arguments. | |||
Our goal is to simplify interoperability in scientific computing by alleviating these difficulties. To this aim, we establish generic open interfaces standards between numerical solvers for typical computational problems that allow to seamlessly interconnect and exchange the models and algorithms implemented by these packages in complex modeling or simulation workflows. Also, we develop a mediator library that connects user code to an implementation code via an interface and automates data marshalling between programming languages so that writing explicit bindings is not required. Therefore, users could switch more easily from one implementation to another, while working through the same programming interface without being concerned with technical details such as calling a function in one programming language from another one. | |||
Specifically, we develop such interfaces for common computational problems such as initial-value problems for ordinary differential equations (time integration), optimization, discrete PDE models based on a reusable language-agnostic core API toolkit. Through a developer platform, we assist other scientific-computing communities with establishing open interface for their respective software stacks. | |||
'''Performance study''' | |||
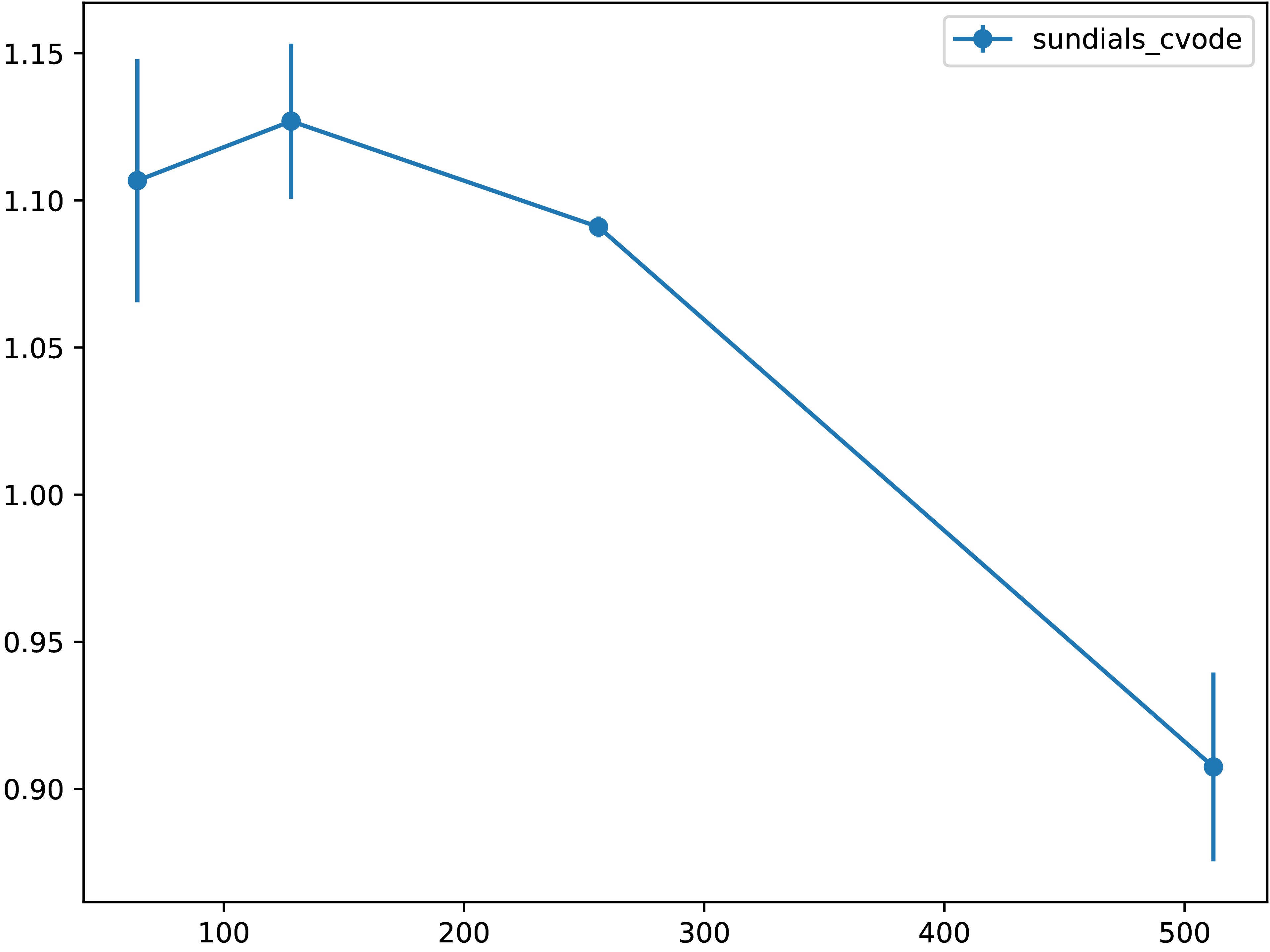
We demonstrate here that the approach described above does not bring a significant performance penalty, as one could expect as a price for flexibility. Precisely, we compare time integration of a two-dimensional reaction--diffusion system via Sundials CVODE solver (which is written in C) invoked from Python directly (i.e., via direct bindings) and via Open Interfaces. As can be seen from the figure below, the performance penalty is only about 10-15 % for very coarse grid resolutions (e. g., 128 × 128), while for moderate grid sizes the performance penalty is negligible. | |||
[[File:Ta2-perf-study-2d-reac-diff-system.png|thumb|center]] | |||
===Model Order Reduction Benchmark (MORB) and its Ontology (MORBO)=== | ===Model Order Reduction Benchmark (MORB) and its Ontology (MORBO)=== | ||
| Line 62: | Line 67: | ||
[https://www.mpi-magdeburg.mpg.de/person/29457/2316 Jan Heiland (MPI Magdeburg)] | [https://www.mpi-magdeburg.mpg.de/person/29457/2316 Jan Heiland (MPI Magdeburg)] | ||
[https://www.uni-muenster.de/AMM/ohlberger/team/hendrik_kleikamp.shtml Hendrik Kleikamp ( | [https://www.uni-muenster.de/AMM/show_perspage.shtml?id=1760 Dmitry Kabanov (University of Münster)] | ||
[https://www.uni-muenster.de/AMM/ohlberger/team/hendrik_kleikamp.shtml Hendrik Kleikamp (University of Münster)] | |||
[https://www.mpi-magdeburg.mpg.de/person/123208/4059055 Kathryn Lund (MPI Magdeburg)] | [https://www.mpi-magdeburg.mpg.de/person/123208/4059055 Kathryn Lund (MPI Magdeburg)] | ||
[https://www.uni-muenster.de/AMM/ohlberger/team/mario_ohlberger.shtml Mario Ohlberger ( | [https://www.uni-muenster.de/AMM/ohlberger/team/mario_ohlberger.shtml Mario Ohlberger (University of Münster)] | ||
[https://www.wwu.de/AMM/ohlberger/team/stephan_rave.shtml Stephan Rave ( | [https://www.wwu.de/AMM/ohlberger/team/stephan_rave.shtml Stephan Rave (University of Münster)] | ||
[https://www.mpi-magdeburg.mpg.de/person/26697/4059055 Jens Saak (MPI Magdeburg)] | [https://www.mpi-magdeburg.mpg.de/person/26697/4059055 Jens Saak (MPI Magdeburg)] | ||
| Line 74: | Line 81: | ||
[https://www.mpi-magdeburg.mpg.de/person/123700/4060519 Pavan Veluvali (MPI Magdeburg)] | [https://www.mpi-magdeburg.mpg.de/person/123700/4060519 Pavan Veluvali (MPI Magdeburg)] | ||
[https://www.uni-muenster.de/AMM/show_perspage.shtml?id=17 Frank Wübbeling ( | [https://www.uni-muenster.de/AMM/show_perspage.shtml?id=17 Frank Wübbeling (University of Münster)] | ||
===Former=== | ===Former=== | ||
| Line 85: | Line 92: | ||
===Interns and Student Assistants=== | ===Interns and Student Assistants=== | ||
Saksham Malhotra (Jun-present 2023) | Saksham Malhotra (Jun-present 2023) | ||
Patrik Pruditsch (Mar-May 2023) | |||
Alexander Stage (Oct-Dec 2022) | Alexander Stage (Oct-Dec 2022) | ||
Latest revision as of 12:15, 26 April 2024
Task area 2: Scientific Computing
Mission
Scientific computing is a cross-disciplinary topic intersecting applied mathematics and computational sciences and engineering (CSE), as well as other scientific areas involving numerical computations, like digital humanities or computational medicine. The principal data types involved are fixed-precision real numbers, which are prone to round-off errors during computation. Beyond the data types found in engineering, such as input/output data of numerical software, in computational mathematics, and specifically in scientific computing, algorithms themselves, as well as their implementations, procedural data, and metadata descriptions, are considered research data.
TA2 will focus on establishing knowledge graphs of numerical algorithms and on building open interfaces for their seamless interconnection in scientific computations. A benchmark framework will employ both to asses performance on a collection of reference data sets from a corresponding area of application while following standardized workflows identified in collaboration with TA4 and other consortia.
Ongoing initiatives
MathAlgoDB (formerly AlgoData)
Our goal is to establish a knowledge graph of numerical algorithms, which interlinks those algorithms with the addressed mathematical problems and associated research data such as journal papers or implementing software packages. As often many variations of essentially the same numerical algorithm are discussed in the literature, an editorial board of domain experts will define the individual algorithm items in the database. The interlinking with the associated data, however, is carried out semi-automatically, using various data sources such as text-mining or metadata search in other databases and MaRDI services.
The measure improves the findability of research data associated with given numerical algorithms and allows non-experts, who need to solve numerical problems in their scientific work, to quickly gain an overview of available methods, their characteristics and implementations. Mathematicians in scientific computing are empowered to easily track the field’s progress by data on competing methods.
We provide an interface for querying and editing data. Note that editing and proposing data is not fully functional yet. The interface comes in a stable version and a beta version which reflects our current working status. The permanent links are mathalgodb.mardi4nfdi.de and main.m1.mardi.ovh.
The beta version is updated daily. Use the deep links below to access Data and the Ontology definition.
| stable | testing | |
|---|---|---|
| Ontology Link | Ontology | Ontology |
| Simple Search | Search | Search |
| Graph Search | Search | Search |
The ontology definition in standard formats (XML, Triples, Turtle) is available under the ontology link above.
At this point, we accept submissions for the graph, respecting the ontology, in standard formats. Please use the definition of the sample Model Order Reduction-Graph as a template. We also support a customized meta format which permits a simpler definition of the graph and has DOI search capabilities for bulk additions to the graph.
Open Interfaces
The scientific-computing community uses several different programming languages for implementing numerical solvers. Moreover, these solvers often have significantly different programming interfaces. Both of these factors impede interoperability, as switching from, say, a Python solver for time integration to another solver implemented in C requires writing bindings and adapting the calling code to different function names and the order of arguments.
Our goal is to simplify interoperability in scientific computing by alleviating these difficulties. To this aim, we establish generic open interfaces standards between numerical solvers for typical computational problems that allow to seamlessly interconnect and exchange the models and algorithms implemented by these packages in complex modeling or simulation workflows. Also, we develop a mediator library that connects user code to an implementation code via an interface and automates data marshalling between programming languages so that writing explicit bindings is not required. Therefore, users could switch more easily from one implementation to another, while working through the same programming interface without being concerned with technical details such as calling a function in one programming language from another one.
Specifically, we develop such interfaces for common computational problems such as initial-value problems for ordinary differential equations (time integration), optimization, discrete PDE models based on a reusable language-agnostic core API toolkit. Through a developer platform, we assist other scientific-computing communities with establishing open interface for their respective software stacks.
Performance study
We demonstrate here that the approach described above does not bring a significant performance penalty, as one could expect as a price for flexibility. Precisely, we compare time integration of a two-dimensional reaction--diffusion system via Sundials CVODE solver (which is written in C) invoked from Python directly (i.e., via direct bindings) and via Open Interfaces. As can be seen from the figure below, the performance penalty is only about 10-15 % for very coarse grid resolutions (e. g., 128 × 128), while for moderate grid sizes the performance penalty is negligible.
Model Order Reduction Benchmark (MORB) and its Ontology (MORBO)
MORB is a demonstrator of the benchmark framework outlined in the MaRDI proposal, and MORBO comprises and ontology and knowledge graph of all the benchmark problem data detailed in the MORWiki. For up-to-date information on MORB and MORBO, visit the corresponding page in the MORWiki.
Description and Design of FAIR CSE Workflows
As a part of this measure we have developed a workflow framework, namely MaRDIFlow, that abstracts the multi-layered components from FAIR computational experiments. Herein, each component will be characterized through an input/output description so that model, code, and data can be used interchangeably and, in the best case, redundantly. Examples and case studies implemented in the MaRDIFlow framework will be updated here soon.
Workshops
First MaRDI Workshop on Scientific Computing, 26-28 October 2022, Münster, Germany
Members
Current
Dmitry Kabanov (University of Münster)
Hendrik Kleikamp (University of Münster)
Mario Ohlberger (University of Münster)
Stephan Rave (University of Münster)
Pavan Veluvali (MPI Magdeburg)
Frank Wübbeling (University of Münster)
Former
Interns and Student Assistants
Saksham Malhotra (Jun-present 2023)
Patrik Pruditsch (Mar-May 2023)
Alexander Stage (Oct-Dec 2022)
Malte Speidel (Mar-May 2022)
Job openings
- Student Assistant at MPI Magdeburg to help with expanding MORBO